Copy number variation in Schizophrenia
 Ever had the feeling you have lost your marbles? According to the Phrase Finder that expression has conveyed a sense of loss, anger, and more recently a lack of common sense or sanity. As it turns out it may be the loss of certain segments of DNA (rather than simple mutations like SNPs) that may have a lot to do with the origins of mental illnesses like schizophrenia.
Ever had the feeling you have lost your marbles? According to the Phrase Finder that expression has conveyed a sense of loss, anger, and more recently a lack of common sense or sanity. As it turns out it may be the loss of certain segments of DNA (rather than simple mutations like SNPs) that may have a lot to do with the origins of mental illnesses like schizophrenia.
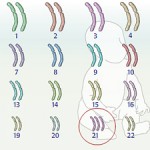
Now before you start thinking that schizophrenics are the only ones to lose their marbles (or large sections of their genomes), It has been previously shown by work like that of Jonathan Sebat of Cold Spring Harbor Laboratory, that deletions, including those larger than a kilobase are common within all of our genomes. Obviously, or not so obviously, most humans seem to get along quite fine with these deletions. However, it has recently been appreciated that many psychiatric disorders seem to be influenced by this genomic structural variation.
PSYCH-CNV’s project aims to look at how copy number variation (CNV) contributes to the development of schizophrenia, amongst other illnesses. In a recent paper entitled Large recurrent microdeletions associated with schizophrenia (Stefansson et.al., Nature 2008), it was hypothesized that rare copy number variations might carry the bulk of the risk. The paper went on to describe three regions where deletions were associated with schizophrenia and related psychoses. PSYC-CNV will focus on rare and de novo variations in order to explain how these unique changes in genome arrangement and organization can explain disorders.
Interestingly, a new method developed by the Sebat lab also aims at increasing our ability to detect these copy number changes and explore their meaning for schizophrenia. Instead of relying on microarrays, which often provide insufficent resolution to detect small CNVs, this is a next generation sequence based approach. Next generation sequencers, such as the Illumina platform used in the above refrenced paper, involve sequencing short reads of DNA which are then assembled into a genome. Sebat and his collaborators at Albert Einstein looked at a paramater called depth of read, instead of just analyzing paired end runs, the previous approach to detecting CNVs within a genome.
| Print article | This entry was posted by Jason Williams on September 1, 2009 at 4:33 pm, and is filed under G2C Online. Follow any responses to this post through RSS 2.0. You can skip to the end and leave a response. Pinging is currently not allowed. |